Medical Out-of-Distribution Analysis Challenge
MICCAI 2023
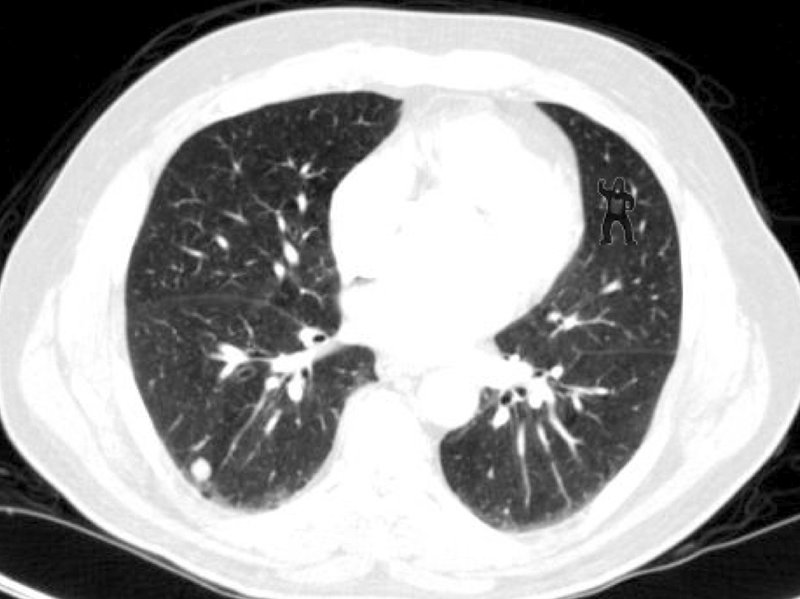
‘Will you be able to spot a Gorilla in a CT scan ???’
Despite overwhelming successes in recent years, progress in the field of biomedical image computing still largely depends on the availability of annotated training examples. This annotation process is often prohibitively expensive because it requires the valuable time of domain experts.
Furthermore, many algorithms used in image analysis are vulnerable to out-of-distribution samples, resulting in wrong and overconfident decisions.
However, even humans are vulnerable to ‘inattentional blindness’. More than 50% of trained radiologists did not notice a gorilla image, rendered into a lung CT scan when assessing lung nodules.
With the Medical Out-of-Distribution Analysis Challenge (MOOD) we want to tackle these challenges!
The MOOD Challenge gives a standardized dataset and benchmark for anomaly detection. We propose two different tasks. First a sample-level (i.e. patients-level) analysis, thus detecting out-of-distribution samples. For example, having a pathological condition or any other condition not seen in the training-set. This can pose a problem to classically supervised algorithms and can further allow physicians to prioritize different patients. Secondly, we propose an object-level analysis i.e. giving a score for each voxel, highlighting abnormal conditions and potentially guiding the physician.
Want more information or take part? Scroll down or visit our Github or Submission-Site.
Data
Datasets
The challenge spans two datasets with more than 500 scans each, one brain MRI-dataset and one abdominal CT-dataset, to allow for a comparison of the generalizability of the approaches. The training set is comprised of hand-selected scans in which no anomalies were identified.
In order to prevent overfitting on the (types of) anomalies existing in our testset, the testset will be kept confidential at all times. As in reality, the types of anomalies should not be known beforehand, to prevent a bias towards certain anomalies in the evaluation. Some scans in the testset do contain no anomalies, whilst others contain naturally occurring anomalies. In addition to the natural anomalies, we will add synthetic anomalies to cover a broad and unpredictable variety of different anomalies and also allow for an analysis of weaknesses and strengths of the methods by different factors.
We believe that this will allow for a controlled and fair comparison of different algorithms.
Number of training samples:
- Brain: 800 scans ( 256 x 256 x 256 )
- Abdominal: 550 scans ( 512 x 512 x 512 )
We provide four toy test-cases for both datasets, for participants to test their final algorithm. After submitting their solution we will report the score on these test-cases back to participants to check if their algorithm run successfully.
To get access to the data go to the Submission-Site.
Details
Tasks
The MOOD Challenge has two tasks:
-
Sample-level: Analyse different scans/samples and report a score for each sample. The algorithm should process a single sample and give a “probability” for this sample being abnormal/ out-of-distribution. The scores must be in [0-1], where 0 indicates no abnormality and 1 indicates the most abnormal input.
In summary: One score per sample.
-
Object-/ Pixel-level: Analyse different scans and report a score for each voxel of the sample. The algorithm should process a single sample and give a “probability” for each voxel being abnormal/ out-of-distribution. The scores must be in [0-1], where 0 indicates no abnormality and 1 indicates the most abnormal input.
The participants can decide if they want to submit a a binary predicition (0 or 1) or a continuous score (0-1). In the case of a continuous score, we use a toy-dataset (similar to the toy-dataset presented to the participants) to determine the best binarization threshold with respect to the presented metric on this toy-dataset. We then use the binarization threshold to binarize the continuous score the binarized score, we apply connected component analysis to find individual objects and discard all objects which are smaller than half the size of the smallest anomaly in our test set and all objects with are more than two times bigger than the biggest object in our test set (to sort out low-frequency noise). This procedure then outputs the prediction objects. We then match the GT objects and the prediction objects in the following way to obtain TP, FP, FN:
If the center of mass of the prediction object falls in the convex hull of the GT object and the size difference is only at max a factor of two (= 0.5 * GT-object size < prediction-object size < 2*GT-object size), then we consider the GT object to be detected, resulting in a TP. If a GT object was not matched/detected by any prediction object we consider this an FN. If a prediction object could not be matched to a GT object, we consider this an FP. We iterate over all images and objects in the dataset and obtain all TP, FP, FN for the whole dataset. The final score is then given by the F1 score (using the determined TPs, FPs, FNs).
In summary: X * Y * Z scores per sample (where X * Y * Z is the dimensionality of the data sample).
Evaluation
- For evaulation please submit a docker for the task on synapse.
- The docker must be able to process a directory with nifti files and…
- … for the Sample-level task output a text file with a single score per sample
- … for the Pixel-level task output a nifti (with the same dimensions as the input) per sample.
- The scores should be in [0-1] (scores above and below the interval will be clamped to [0-1]).
- If a case is missing or fails we will assign it the lowest given anomaly score (= 0).
- During the evaluation, a runtime of 600 sec/case is allowed (You will get a report of the runtime along with your toy example scores).
- No internet connection will be available during the evaluation.
- Only the provided output directory will have write access.
- We will use the reported scores together with the ground truth labels to calculate the AP over the whole dataset (for more information regarding the AP see scikit-learn ).
- We combine the two datasets by choosing a consolidation ranking schema.
- Teams are allowed 3 submissions, however, only the latest submission will be considered (due to technical issues we increased the #submissions ).
To see our evaluation code and reproduce the results on the toy cases visit our github-page.
Rules
- Only fully automatic algorithms allowed.
- It is only allowed to use the provided training data. No other data and data sources are allowed.
- The teams may decide after receiving their challenge rank if only their username or teamname including the team members will appear on the leaderboard.
- By default (i.e. if they don’t decline) the winning team will be announced publicly. The remaining teams may decide if they choose to appear on the public Leaderboard.
- Teams that reveal their identity, can nominate members of their team as co-author for the challenge paper.
- We reserve the right to exclude teams and team members if they do not adhere to the challenge rules and guidelines.
- The method description submitted by the authors may be used in the publication of the challenge results. Personal data of the authors can include their name, affiliation and contact addresses.
- Participating teams may publish their own results separately.
- Teams are allowed three submissions, however, only the latest submission will be considered.
- All participating teams can choose if they will appear on the leaderboard. However (to prevent misconduct) only teams that open source their code will be eligible to win the challenge and receive any prizes.
schedule
Event Program:
12 Oct 2023
| 13:30 - 13:35 PDT | Welcome |
| 13:35 - 14:05 PDT | Keynote: Latha Pemula (Applied Scientist, AWS AI Labs, Homepage) |
| Challenge Intro | 14:05 - 14:15 PDT |
| 14:15 - 15:45 PDT | Participants Presentations |
| 15:45 - 16:00 PDT | Coffee Break |
| 16:00 - 16:30 PDT | Results |
Important Dates:
| Challenge Data Release: | 01 April 2023 |
| Registration Until: | 30 August 2023 |
| Submission of Dockers closes: | |
| Submit abstract before: | |
| Announcement of Results: | Miccai 2023 |
leaderboard
Sample-Level:
| Rank | Team | Further Info |
|---|---|---|
| 1 | SubmitSomething | Publication & GitHub |
| 2 | AIRI | |
| 3 | CISTIB | Paper |
| 4 | TUe IMAGe | |
| 4 | IntGeeks | |
| 6 | LucasGago | |
| 7 | BCTS |
Pixel-Level:
| Rank | Team | Further Info |
|---|---|---|
| 1 | SubmitSomething | Publication & GitHub |
| 2 | TUe IMAGe | Paper |
| 3 | IntGeeks | |
| 4 | BCTS |
You can find the presentation of the results here.
how-to
Register for the Challenge
Go to <synapse.org> and click on Join.
Get access to the data
Go to <synapse.org> and go to the Data tab. In the mood folder, you can access the data.
We suggest the following folder structure (to work with our github examples):
mood/
--- brain/
------ brain_train/
------ toy/
------ toy_label/
--- abdom/
------ abdom_train/
------ toy/
------ toy_label/Quick-Start
Join the Challenge, download the Data and have a look at our ready-to-go examples or last year’s submissions http://medicalood.dkfz.de/web/2020/#leaderboard.
Load the scans
In python you can load and write the nifti files using nibabel:
-
Install nibabel:
pip install nibabel -
Load the image data and affine matrix:
1
2
3
4
5
import nibabel as nib
nifti = nib.load(source_file)
data_array = nifti.get_fdata()
affine_matrix = nifti.affine
- Save a array as nifti file:
1
2
3
4
import nibabel as nib
new_nifti = nib.Nifti1Image(data_array, affine=affine_matrix)
nib.save(new_nifti, target_file)
Build a docker
1. Requirements
Please install and use docker for submission: https://www.docker.com/get-started
You can build and use any docker base/ image you like. There are already good base docker images to build on for:
- Pytorch: https://hub.docker.com/r/pytorch/pytorch/
- Tensorflow https://www.tensorflow.org/install/docker
For GPU support you may need to install the NVIDIA Container Toolkit: https://github.com/NVIDIA/nvidia-docker
2. Docker Setup
For the different tasks the docker needs the following scripts (which accept parameter input_folder and output_folder):
-
Sample-level:
/workspace/run_sample_brain.sh input_folder output_folder(for the brain dataset)
/workspace/run_sample_abdom.sh input_folder output_folder(for the abdominal dataset) -
Pixel-level:
/workspace/run_pixel_brain.sh input_folder output_folder(for the brain dataset)
/workspace/run_pixel_abdom.sh input_folder output_folder(for the abdominal dataset)
The docker has to allow mounting the input folder to /mnt/data and the output folder to /mnt/pred. We will mount the input and output folder and pass them to the run scripts.
You will only have write access to /mnt/pred so please also use it for temporary results if needed.
During testing, the docker image will be run with the following commands:
- Sample-level:
docker run --gpus all -v "input_dir/:/mnt/data" -v "output_dir:/mnt/pred" --read-only docker-image-name /workspace/run_sample_brain.sh /mnt/data /mnt/pred
docker run --gpus all -v "input_dir/:/mnt/data" -v "output_dir:/mnt/pred" --read-only docker-image-name /workspace/run_sample_abdom.sh /mnt/data /mnt/pred
- Pixel-level:
docker run --gpus all -v "input_dir/:/mnt/data" -v "output_dir:/mnt/pred" --read-only docker-image-name /workspace/run_pixel_brain.sh /mnt/data /mnt/pred
docker run --gpus all -v "input_dir/:/mnt/data" -v "output_dir:/mnt/pred" --read-only docker-image-name /workspace/run_pixel_abdom.sh /mnt/data /mnt/pred
Your model should process all scans and write the outputs to the given output directory.
Please be aware of the different output formats depending on your task:
- Sample-level: For each input file (e.g. input1.nii.gz) create a txt file with the same name and an appended “.txt” file-ending (e.g. input1.nii.gz.txt) in the output directory and write out a single (float) score.
- Pixel-level: For each input file (e.g. input1.nii.gz) create a nifti file with the same dimensions and save it under the same (e.g. input1.nii.gz) in the output directory.
For more information have a look at our github example: https://github.com/MIC-DKFZ/mood
Test a docker locally
You can test you docker locally using the toy cases. After submitting your docker, we will also report the toy-test scores on the toy examples back to you, so you can check if your submission was successful and the scores match.
To run the docker locally you can:
- Either run the docker manually and use the
evalresults.pyfile (not recommended) - Or use the
test_docker.pyscript (recommended):
Clone the github repo: git clone https://github.com/MIC-DKFZ/mood.git
Install the requirements: pip install -r requirements.txt
Run the test_docker.py script:
python test_docker.py -d docker_image_name -i /path/to/mood -t samplewhere with -d you can pass your docker image name, with -i you pass the path to your base input directory which must contain a brain and an abdom folder (see above -> ‘Get access to the data’), and with -t you can define the task, either sample or pixel.
Test a docker on our submission system
You can test you docker on our submission system only for the toy cases. After submitting your docker, we will report the toy-test scores on the toy examples back to you, so you can check if your docker runs successful and the scores match with your local system.
To test your docker please submit to the tasks Toy Examples - Pixel-Level or Toy Examples - Sample-Level.
In the next Section on “Submit a docker” you can see how to submit a docker but submit to the tasks Toy Examples - Pixel-Level or Toy Examples - Sample-Level only.
Please not that this will not count as a MOOD 2020 submission. Submitting to the Toy Examples tasks will not increase you submission count and will not include your submission in the challenge.
Submit a docker
-
Becoming a certified Synapse user:
Important: In order to use all docker functionality you must be a certified user : https://www.synapse.org/#!Quiz: -
Create a new project on synapse:
-
To submit a docker file, you first need to create a new project on the synapse platform (e.g:
MOOD_submission_<task>_<Your team name>). -
Note the Synapse Project ID (e.g. syn20482334).
-
The organizing team of the challenge (Medical Out of Distribution Analysis Challenge 2020 Organizers) must be given download permissions to your project (Under Project Settings -> Project Sharing Settings ),
-
-
Upload docker-image to synapse:
1
2
3
docker login docker.synapse.org
docker tag docker_imagename docker.synapse.org/synapse_project_ID/docker_imagename
docker push docker.synapse.org/synapse_project_ID/docker_imagename:latest
- Submit the docker image:
- Got to your Project Page → Docker and click on the docker file .
- Click on Docker Repository Tools → Submit Docker Repository to Challenge .
- Choose the ‘tag’ you want to submit.
- Choose the task you want to submit to (either ‘Sample-level’ or ‘Pixel-level’) (if you plan to participate in multiple tasks of the challenge, you need to submit your corresponding docker to each queue individually) .
- Specify if you are entering alone or as a team.
Common Errors
-
Read-only files system:
OSError: [Errno 30] Read-only file system: '/workspace/XXXXPlease make sure only to write your (temp) data to
/mnt/predand perhaps setENV TMPDIR=/mnt/pred. -
Python site packages permission error:
File "/opt/venv/lib/python3.X/site-packages/pkg_resources/__init__.py", line XXXX, in get_metadata with io.open(self.path, encoding='utf-8') as f: PermissionError: [Errno 13] Permission deniedPlease make sure you use virtual environments and use the virtualenv path as the first place to check for packages.
-
Relative Paths:
FileNotFoundError: [Errno 2] No such file or directory: './XXX'Please only use absolute paths, i.e.
/workspace/XXX. -
Any other Errors ? Feel free to contact us =).
Submit the abstract
To offically take part in the challenge, the participants are required to provide a short abstract describing their submission. You can find the abstract templates on Synapse or Overleaf. Please fill out the provided template and send it as pdf to d.zimmerer@dkfz.de before the 07 September 2022.
Cite the Challenge
Please be sure to include the following citations in your work if you use the challenge data:
@article{zimmerer2022mood,
title={MOOD 2020: A public Benchmark for Out-of-Distribution Detection and Localization on medical Images},
author={Zimmerer, David and Full, Peter M and Isensee, Fabian and J{\"a}ger, Paul and Adler, Tim and Petersen, Jens and K{\"o}hler, Gregor and Ross, Tobias and Reinke, Annika and Kascenas, Antanas and others},
journal={IEEE Transactions on Medical Imaging},
volume={41},
number={10},
pages={2728--2738},
year={2022},
publisher={IEEE}
} For more information see https://zenodo.org/record/6362313.
More
Organizers:
David Zimmerer, Jens Petersen, Gregor Köhler, Paul Jäger, Peter Full, Klaus Maier-Hein
Div. Medical Image Computing (MIC), German Cancer Research Center (DKFZ)
Tobias Roß, Tim Adler, Annika Reinke, Lena Maier-Hein
Div Computer Assisted Medical Interventions (CAMI), German Cancer Research Center (DKFZ)
Links:
- Offical Website/ Submission system: https://synapse.org/mood
- Github-Repository: https://github.com/MIC-DKFZ/mood
- Challenge-Document: https://zenodo.org/record/6362313
- Miccai 2022: https://conferences.miccai.org/2023/en/
- Some Anomaly Detection/ OoD papers: https://paperswithcode.com/task/anomaly-detection , https://paperswithcode.com/task/out-of-distribution-detection
- Previous editions: http://medicalood.dkfz.de/web/2020/ , http://medicalood.dkfz.de/web/2021/, http://medicalood.dkfz.de/web/2022/
- Inattentional blindness in expert observers paper: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3964612/
- NVidia: https://www.nvidia.com/
- Theme: https://github.com/t413/SinglePaged